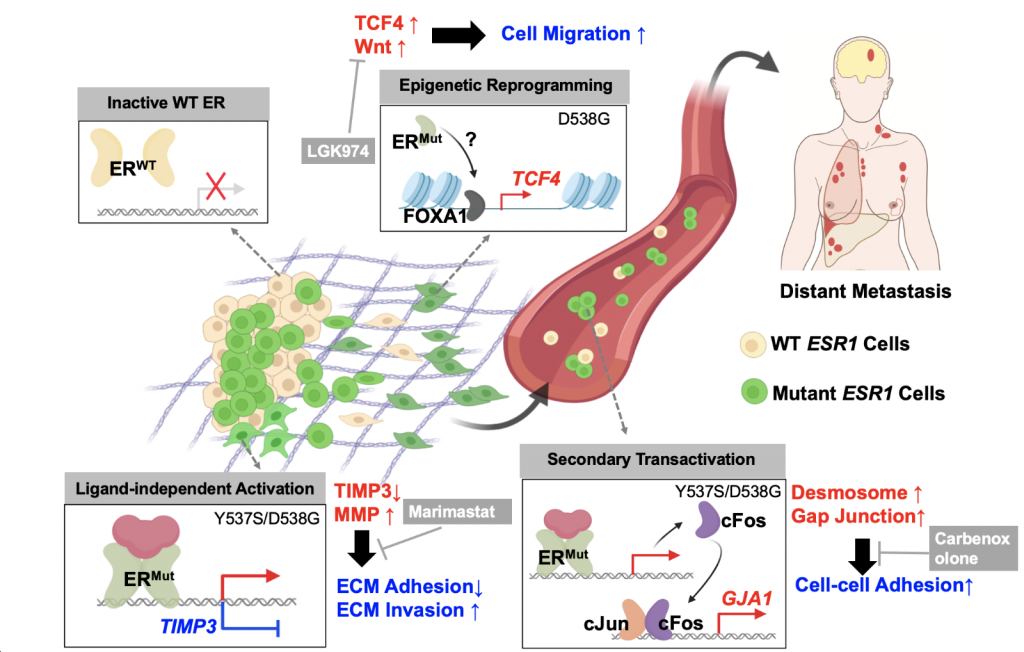
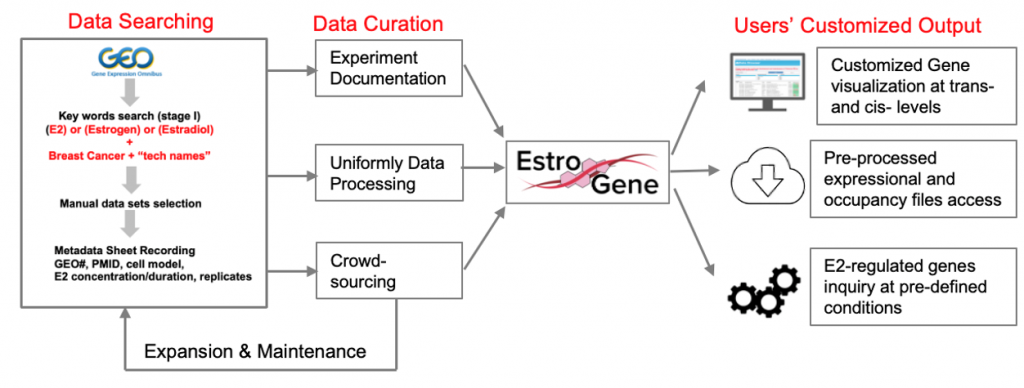
We are pleased to introduce the EstroGene Project -a comprehensive multi-omic NGS database focusing on estrogen receptor function in breast cancer. It aims to document and integrate the majority of publicly available estrogen-stimulated next generation sequencing data sets (including RNA-seq, microarray, ChIP-seq, ATAC-seq, ChIA-PET, Hi-C, GRO-seq, etc), and establish a comprehensive database to allow users’ customized data mining and visualization. We have curated 136 published NGS data sets from 2004-2022 across 19 breast cancer cell lines and generated a browser for simplified queries.
Features of EstroGene:
-A uniformly processed and crowd-sourced multi-omic database with detailed experimental documentation summary.
-A browser allowing single gene-based visualization of E2-induced expressional changes and ER proximal binding at users’ selected genes of interest.
-A browser supporting statistical cutoff-based gene list query function to export genes regulated by E2 under users’ defined contexts.
-An ER and breast cancer-centered database for dissecting the biological and technical diversity and variation of estrogen receptor-relevant NGS experiments and the confound ER regulomes in breast cancer.
We have summarized all of the curated datasets in this google form. We are crowd-sourcing additional datasets that may not be available in the public domain but are available within laboratories. If you have such a dataset please don’t hesitate to fill in the google form and we will contact you back.
We would appreciate it if you could operate the website and give us feedbacks to improve it and continue notify us about new data sets via the google form.
The BioRxiv manuscript of this project will be deposit after receiving feedbacks from the research community!
For any queries please email Nadine Ryan (ryann@upmc.edu)