


Julia Foldi MD and colleagues from the Lee-Oesterreich laboratory collaborated with Natera Inc to report on retrospective analysis of real-world data in 66 patients with metastatic ILC using a clinically validated, personalized, tumor-informed ctDNA assay (Signatera). Serial ctDNA testing in patients with mILC is feasible and may enable personalized surveillance and real-time therapeutic monitoring.
https://pubmed.ncbi.nlm.nih.gov/40324116

Members of the lab attended this year’s ILC symposium in Leuven, Belgium.































Congrats to Neil, Adrian, and Steffi on their new publication in JAMA Surgery! They led a nonrandomized trial evaluating a “nudge” intervention embedded in the electronic health record to decrease use of low-value axillary surgery for older patients with ER+ breast cancer. The nudge was widely successful and decreased use of SLNB by about 50% over the study period. This study included a strong collaborative component with researchers from breast surgical oncology, general internal medicine, statisticians, and AI/ML experts. Full link to the paper can be found here:

https://jamanetwork.com/journals/jamasurgery/fullarticle/2821213
Adrian discusses the latest in transcriptomics shedding light on cancer heterogeneity and its impact on treatment strategies.
@UPMC#SABCS22#BreastCancer Listen and Share the Audio Podcast Here: https://oncologytube.com/v/41787
We are pleased to introduce the EstroGene Project -a comprehensive multi-omic NGS database focusing on estrogen receptor function in breast cancer. It aims to document and integrate the majority of publicly available estrogen-stimulated next generation sequencing data sets (including RNA-seq, microarray, ChIP-seq, ATAC-seq, ChIA-PET, Hi-C, GRO-seq, etc), and establish a comprehensive database to allow users’ customized data mining and visualization. We have curated 136 published NGS data sets from 2004-2022 across 19 breast cancer cell lines and generated a browser for simplified queries.
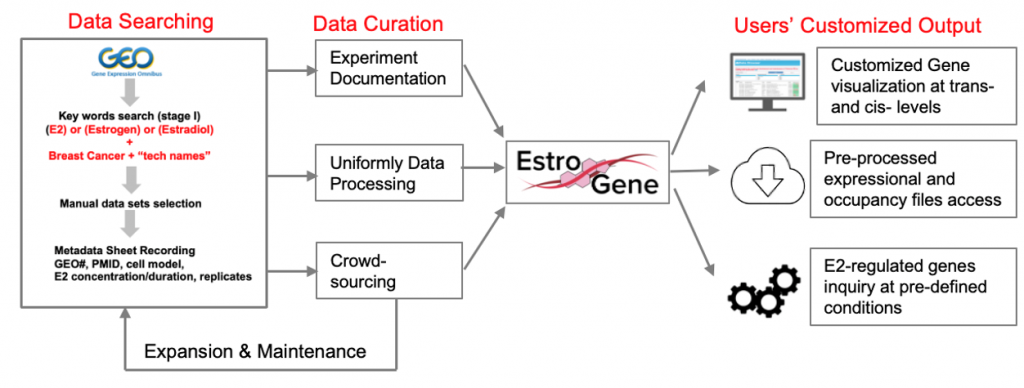
Features of EstroGene:
-A uniformly processed and crowd-sourced multi-omic database with detailed experimental documentation summary.
-A browser allowing single gene-based visualization of E2-induced expressional changes and ER proximal binding at users’ selected genes of interest.
-A browser supporting statistical cutoff-based gene list query function to export genes regulated by E2 under users’ defined contexts.
-An ER and breast cancer-centered database for dissecting the biological and technical diversity and variation of estrogen receptor-relevant NGS experiments and the confound ER regulomes in breast cancer.
We have summarized all of the curated datasets in this google form. We are crowd-sourcing additional datasets that may not be available in the public domain but are available within laboratories. If you have such a dataset please don’t hesitate to fill in the google form and we will contact you back.
We would appreciate it if you could operate the website and give us feedbacks to improve it and continue notify us about new data sets via the google form.
The BioRxiv manuscript of this project will be deposit after receiving feedbacks from the research community!
For any queries please email Nadine Ryan (ryann@upmc.edu)
Research publication: Clinicopathological Features and Outcomes Comparing Patients With Invasive Ductal and Lobular Breast Cancer

Multi-institutional study of #lobular cancer led by my long-term collaborator @oesterreichs. ILC has worse long-term outcomes than IDC – lots more research to be done. @LobularIreland @LobularBCUK @LobularBCA @LobularResearch @BCRFcure @SusanGKomen #bcsm pic.twitter.com/4tXz7UnOtd
— Adrian Lee (@Adrianvlee) October 16, 2022

Congrats to lab member Neil Carleton and surgeon / lab collaborator Dr. Priscilla McAuliffe on their recent publication in Nature Reviews Clinical Oncology! This comment explores how chronological age cutoffs in clinical oncology guidelines are defined. Using the case of breast cancer in older women, the authors discuss why the age at which individuals transition from ‘younger’ to ‘older’ has been defined in a heterogeneous, unstandardized, arbitrary and disparate manner.
Read the full article here: https://www.nature.com/articles/s41571-022-00684-4.


This study was led by former graduate student Vaciry Li with great efforts from many intra- and inter-group collaborations. Congratulations to all!
In this study, we showed that context and allele-dependent transcriptome and cistrome reprogramming in ESR1 mutation cell models, which elicit diverse metastatic phenotypes related to cell-cell adhesion, cell-ECM adhesion and migration driven by increased desmosome/gap junctions, dampened TIMP3-MMP axis and Wnt pathway. Importantly, some of these pathways can pharmacologically targeted and reveals novel therapeutic strategies.
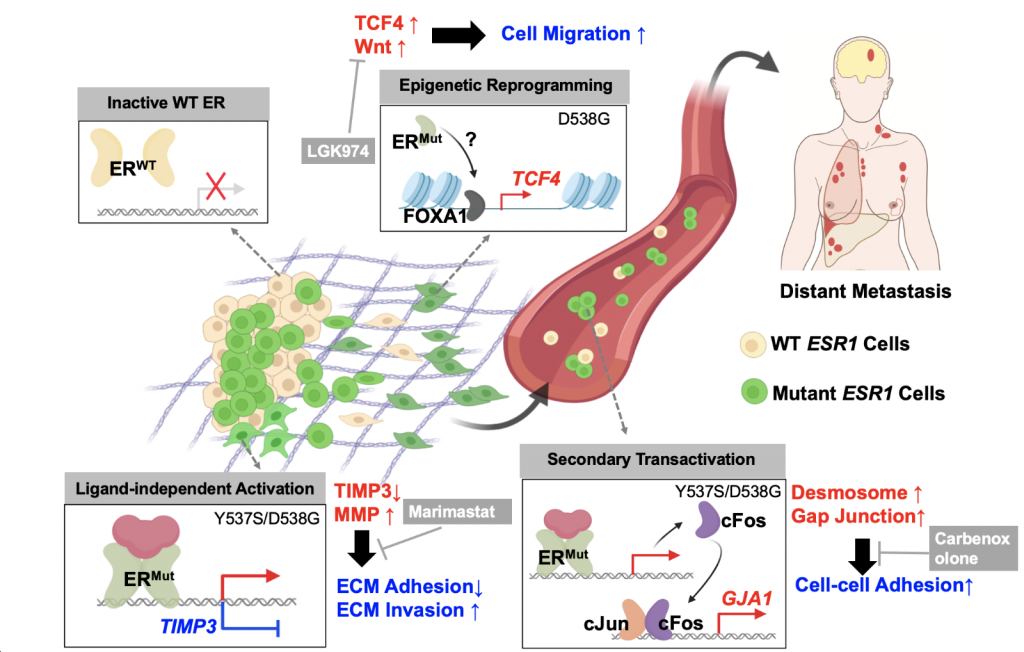
Our paper is out on ESTROGEN RECEPTOR MUTATIONS – so much more than endocrine resistance. Multiple roles in #metastaticbreastcancer Great work by @ZheqiL @UPMCHillmanCC @MageeWomens now postdoc @DanaFarber @Adrianvlee https://t.co/X1baEXelai pic.twitter.com/nI7VNbrzau
— Dr. Steffi Oesterreich (@oesterreichs) January 27, 2022